Recently, the subcellular localization and dynamics of RNA have been attracting attention as being involved in various cellular functions. We have developed a technology to selectively bind proteins to arbitrary RNA 8 nucleotide sequences using the RNA-binding protein domain of PUM-HD.
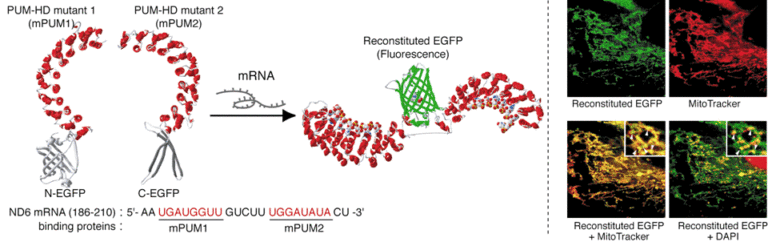
Using this technology with the split fluorescent protein reconstitution method, we have constructed an RNA probe that emits light when it binds to the target RNA and functions in living cells. Widely used RNA probes have various limitations, such as the need for chemical fixation of cells, low signal-to-background ratios, or the need to fuse large tag sequences to the target RNA. Our technology can label and visualize endogenous RNA that has not been artificially processed by simply introducing a gene vector encoding the probe into the cell. In addition, since fluorescence is restored only after the probe molecule binds to the target RNA, the signal-to-background ratio is high and can be used for single molecule fluorescence observation. The developed probe and labeling technology can be applied to various RNAs in living cells. Using this probe, we have observed NADH dehydrogenase mRNA in mitochondria, β-actin mRNA in cytoplasm, and telomere repeat sequence containing RNA (TERRA) in the nucleus. The localization and transport of intracellular RNAs are closely related to the expression of various cellular functions such as cell migration, development, and differentiation, etc. The RNA labeling and visualization technique using PUM-HD is expected to contribute to the elucidation of the mechanisms of different cellular physiological functions.

- T. Yamada, H. Yoshimura, R. Shimada, M. Hattori, M. Eguchi, T. K. Fujiwara, A. Kusumi, T. Ozawa, Sci. Rep. 6, 38910 (2016). DOI: 10.1038/srep38910
- H. Yoshimura, A. Inaguma, T. Yamada and T. Ozawa, ACS Chem. Biol., 7, 999-1005 (2012). DOI: 10.1021/cb200474a
- T. Ozawa, Y. Natori, M. Sato, Y. Umezawa, Nat. Methods. 4, 413–419 (2007). DOI:10.1038/nmeth1030
